Human blood groups are interesting from both medical and evolutionary perspectives. Different blood groups confer resistance or susceptibility to a wide range of infectious disease and, likely as a result of this, have been under long-term balancing selection across primates. They also vary dramatically in frequency across human populations and, probably, across time as well. Here, we use ancient DNA to estimate the frequencies of the two most commonly discussed groups - specifically the ABO and Rhesus systems - in Europe over the past ten thousand years. We show that some ancient populations of Europe had blood group frequencies that were more extreme than any present-day population. In particular, Mesolithic hunter-gatherers had a higher frequency of type O than any present-day population, and both hunter-gatherers and bronze-age steppe populations had a higher frequency of the Rhesus negative allele than any present-day population.
ABO
The ABO blood group system was the earliest to be discovered. ABO blood group is determined by polymorphisms in the ABO gene. In particular, the type O allele is determined by a 1 base deletion (rs8176719) and if that deletion is not present then type A and B are determined by the SNPs rs8176746 and rs8176747. Actually it is more common than that, for example there are many other loss-of-function mutations that lead to the O phenotype, but these are relatively rare.
In evolutionary sense, types A and B are the oldest, since those polymorphisms are shared with other primates. The human type O allele is a more recent mutation, but still probably at least one million years old (the Altai Neanderthal is type O, for example). In present-day populations, all three alleles segregate in Africa, A/O are most common in Europe with B~5-10%, B is relatively common in Asia, but O is virtually fixed in Native Americans.
Rhesus
The Rhesus blood group system is the second most commonly encountered system, determined by polymorphism at the RHD gene. Again, this system has a large number of different groups, but the most important is the Rh- group, where the gene is completely inactivated. In Europe, the most common such allele is actually a deletion at chr1:25592642-25661222, which completely deletes RHD. This polymorphism is at around 10-20% frequency in African populations, very rare in East Asia, and relatively common (~40%) in Europe. The allele is recessive, so you need two copies of the Rh- allele to have the Rh- phenotype.
This high frequency in Europe has long been surprising because the Rh- phenotype has an obviously deleterious effect. In particular, if an Rh- mother has an Rh+ child (because the child inherited a Rh+ allele from its father), then it’s possible for the mother to produce antibodies against the Rh+ antigen leading to haemolytic disease and severe illness for the child. There are three common explanations for the high frequency of the Rh- allele. First, there might be some (unknown) beneficial effect of the Rh-allele. Effects like this related to malaria resistance are what drive the high frequencies of the sickle cell trait and many thalassemias. On the other hand, no obvious selective advantage is known, there are no obvious genomic signals of selection on the Rh- allele, and if there were a selective advantage to the Rh- allele then we might ask why it hasn’t fixed, since once the Rh- frequency rose above 50%, it would be selected for, rather than against. So this only really works if it is overdominant. Another possible explanation is reproductive overcompensation - Rh- women have more children to replace the ones that die for haemolytic disease. But this seems unlikely. The range of parameters for which this model works is relatively small, particularly since the effect gets worse for subsequent pregnancies. A third explanation, suggested by Haldane in 1942 is that the high frequency in Europe is due to the fact that present-day Europeans are recently admixed between a population that has a very high frequency of the Rh- allele and one that a very low frequency. In fact, the ancient DNA evidence suggests that something like this is close to the truth. Cavalli-Sforza and colleagues thought that this was specifically a mixture between Rh- hunter-gatherers and Rh+ Farmers, largely based on the observation that the Basque population, who they believed to have largely hunter-gatherer ancestry, have a very high frequency of the Rh- phenotype.
Ancient DNA
I looked at these alleles in ancient samples dating to the past 12,000 years, all from West Eurasia. I divided them into four groups:
- Hunter-gatherers (HG); Mesolithic and neolithic individuals with ancestry that is like that of Mesolithic Europeans.
- Early Farmers (EF); Neolithic individuals from NW Anatolia, and European Neolithic individuals with similar ancestry, plus small (~0-20%) amounts of hunter-gatherer ancestry.
- Steppe (ST); Bronze Age individuals with “Yamnaya-like” ancestry
- Bronze Age (BA); Individuals from Bronze Age Europe that have a mixture of the three other types of ancient ancestry.
I also compared with present-day European populations, separating out Finns (FIN) and Non-Finns (NFE) (Finns have more steppe-like ancestry).
The ABO determining variants are on the 1240k capture array that has been used to genotype many samples, so I was able to look them up in a sample of around ~500 ancient individuals (the same individuals used here). I looked up present-day allele frequencies in gnomAD. It’s harder to genotype the RHD deletion from capture data so I used a smaller dataset of around ~200 individuals with published shotgun data to genotype this deletion. I couldn’t find it in gnomAD so I used allele frequencies from 1000 Genomes.
it tuns out that the O allele is at high frequency in hunter-gatherers, but relatively rare on the Steppe. The B allele seems to be absent in both hunter-gatherers and early farmers, and seems to be introduced from the steppe in the Bronze Age. The Rh- allele seems to be relatively common in hunter-gatherers and, particularly, in steppe populations, and relatively rare in early farmers, partly confirming Haldane and Cavalli-Sforza’s hypotheses. Allele frequency estimates are in the figures below (bars show 95% binomial confidence intervals).
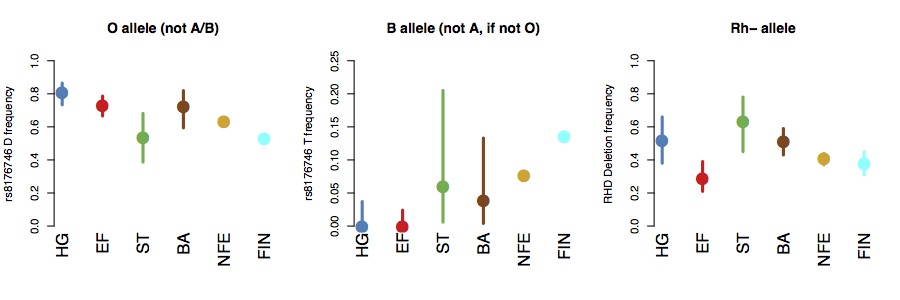
If we compute expected phenotypic frequencies, this suggests that around around 65% of Mesolithic hunter-gatherers would have been type O, compared to around 40% in present-day Europeans, and around 40% of Steppe-ancestry individuals would have been Rh-, compared to around 24% of hunter-gatherers, 4% of early farmers, and about 16% of present-day Europeans.
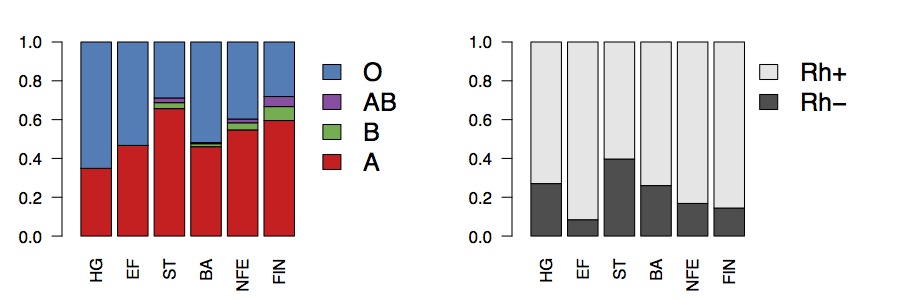
Broadly speaking, the present-day frequencies are consistent with genome-wide ancestry, in the sense that they are just a mixture of the ancient populations (roughly equal to the Bronze Age). The Rh- frequency in present-day Europe is a bit lower than in the Bronze Age, which might be evidence for selection against the allele, but might also just be because there’s a bit less Steppe ancestry in 1000 Genomes than in the samples Bronze Age populations. This doesn’t really explain the Basque frequency though. The Basque population doesn’t particularly have a lot of hunter-gatherer or Steppe ancestry. But perhaps there is substructure in Rh- frequencies within the Basque population, or within the hunter-gatherer populations. Finally, we’ve explained the present-day frequency in terms of mixtures of ancestral populations, but really we have just pushed the question back ten thousand years. Why was the Rh- frequency so high in hunter-gatherer and Steppe populations? Clearly there has to be some selective advantage to this allele at some point, otherwise it would just have been removed everywhere. Perhaps in these populations that selective advantage was stronger. Or perhaps they were just sufficiently small that it was able to drift to high frequency.