The lactase persistence allele
In European ancestry populations, the LCT/MCM6 locus on chromosome 2 exhibits one of the strongest signals of a hard selective sweep in the entire genome (Grossman et al. 2013, Mathieson et al. 2015). This is assumed to be due to selection on the derived A allele of rs4988235 (i.e. the T allele of -13910 C>T) which is associated with adult lactase persistence (Ennatah et al. 2002), allowing the digestion of lactose—and thus unfermented milk—into adulthood. This is a plausible hypothesis, particularly since different mutations with similar phenotypic effects have arisen independently several times (e.g. Ranciaro et al. 2014). However, the reason for selection remains somewhat unclear and none of the proposed hypotheses (access to energy content, hygiene, vitamin D or calcium, etc… ) seem completely convincing (See Szpak et al. 2019 for example).
Observations from ancient DNA
Ancient DNA has shown that the persistence mutation did not, as one might have assumed, arrive in Europe with the first farmers and actually only became common in the Bronze Age, many millenia after the domestication of cattle and the start of dairying (Burger et al. 2007, Mathieson et al. 2015, Mathieson & Mathieson 2018). With the large aDNA datasets now available, we are able to track the spread of the allele, at least in some parts of Western Eurasia with very high temoral and spatial accuracy. In principle, this allows us to put constraints on the time and place at which selection operated, and perhaps to support or disprove some of the hypothesis about the drivers of selection.
I collected published data from 1917 ancient West Eurasians (mostly 1240K capture data), of which 774 had coverage at rs4988235. I also collected present-day alelle frequencies from Liebert et al. 2017. In the maps below, light/dark blue dots represent ancestral/derived alleles.
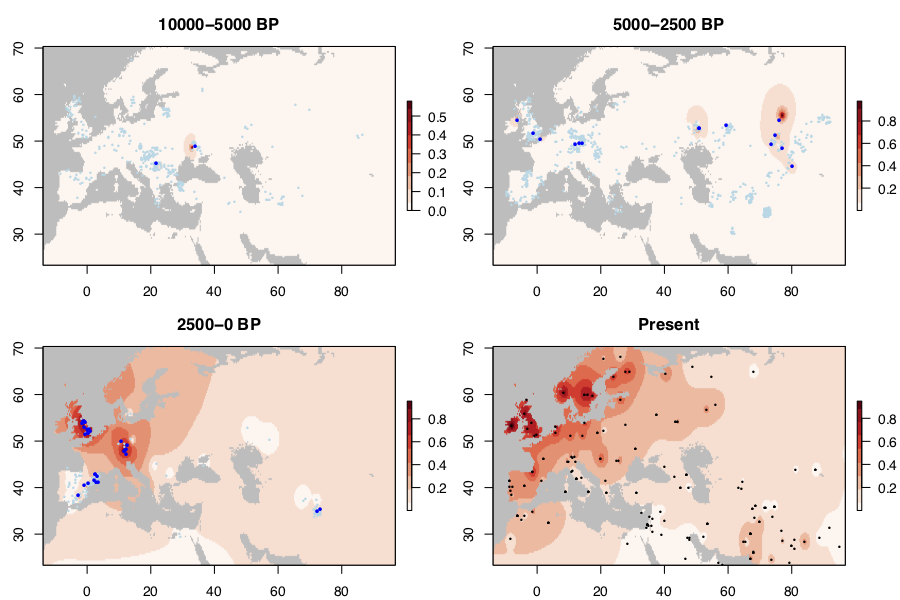
Before 5000 BP, there are only a couple of occurrences of the allele. It’s quite possible that these are genotyping or dating errors. By 2500 BP, the allele is present over a band stretching from Ireland to Central Asia at around 50 degrees latitude. This probably reflects the spread of Steppe ancestry populations in which the allele originated. However, the allele is still rare (say <1% frequency) over this entire range. It does not become common anywhere until some time in the past 2500 years - when it reaches its present-day high frequency in Britain and Central Europe. It’s also at high frequency today in Scandinavia. I don’t have any ancient DNA from the past 2500 years from the region, but Margaryan et al. 2019 suggests that the trajectory was very similar to Britain and Ireland. The allele is relatively rare in Iberia in this period, but intermediate frequency today (45% frequency in 1000 Genomes IBS).
Timing and location of selection
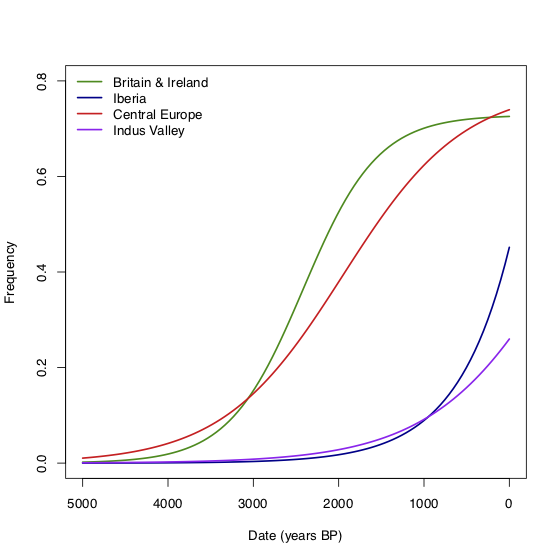
Since we have relatively dense sampling in Britain & Ireland, Central Europe and Iberia (and a few samples in the Indus valley), we can look at the trajectory of the allele in each of these regions separately. The figure below shows a logistic growth model fitted to the ancient and modern (from 1000 Genomes) data in these regions. In Britain, the allele doesn’t start to increase in frequency until perhaps 3000-4000 BP. It then increases rapidly until around 1500 BP, when it seems to level off at around the present-day frequency. There’s a similar pattern in Central Europe, consistent with measurements of the allele frequency in Medieval Germany (Kruttli et al. 2014). In contrast, in Iberia, the increase starts much later—perhaps 1000-2000 BP—but doesn’t obviously level off. What does this tell us about the things that might be driving selection? It’s possible that the start time of the increase is limited by the absence of the allele, rather than the absence of selection. But if selection really stops at 1500 BP, then it would suggest that whatever is driving selection stops in Britain, Ireland and Central Europe at that time, but is still present in Iberia. Another possibility might be some kind of frequency-dependent selection although I have no idea how that would work in practice.
South Asia
One of the interesting things about the “European” persistence allele is that it is also relatively common in parts of South Asia (Gallego Romero et al. 2012). This is likely because the same haplotype was brought to South Asia through gene flow with relatives of the same Steppe populations that brought it to Europe. Looking at the data from Narasimhan et al. 2019 it seems that the allele appeared in South Asia much later than in Europe. Sampling is a bit limited, but its earliest appearance in data is around 2000 BP in Butkara and Swat. The present-day frequency of ~25% in some South Asian populations (e.g. 1000 Genomes PJL) suggests strong, recent selection perhaps similar to what we see in Iberia. In any case seems like selection on the alelle was in parallel in Europe and South Asia. It is probably not the case that the Steppe ancestry in South Asia was from a population that already had a high frequency of the allele.