What is the probability that a site that is variable in modern humans is also variable in archaic humans? This question followed on from some discussion on Twitter about how unusual it was to find a derived allele shared between modern and archaic humans. I recently looked at this empirically with the 1000 Genomes data. The short answer is that if a derived allele is at high frequency in present-day African populations, then it is quite likely to be present in Archaic humans due to incomplete lineage sorting (i.e. it was polymorphic in the common ancestors of modern and archaic humans). If it’s at high frequency in non-Africans but rare in Africans, then it is quite likely to be present in archaic humans but, presumably, to have introgressed into modern humans.
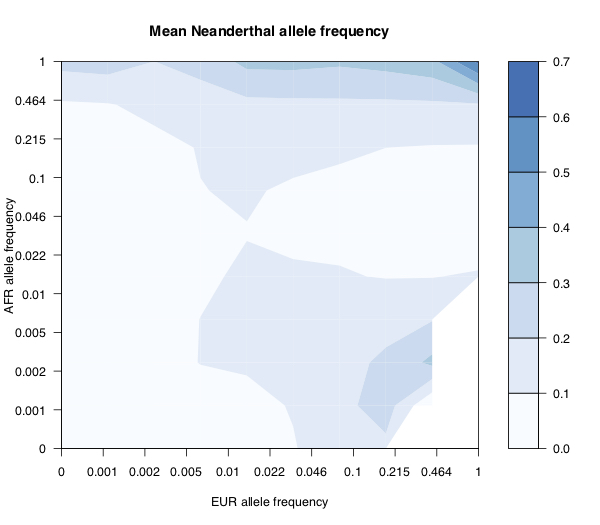
The plot above shows the mean Neanderthal derived allele frequency (shown by colour), as a function of the European and African super-population derived allele frequencies (log scaled), for 1000 Genomes SNPs. Along the top of the plot, you see that anything that’s common (say >50% frequency) in African populations has relatively high expected frequency in Neanderthals. Towards the bottom right, you see that variants that are common in Europe but rare in Africa are also at relatively high frequency in Neanderthals - presumably these are introgressed. Compare with the same plot for Denisovans (below). As before, anything that’s common in Africa is likely to be present in Denisovans, but there’s no sign of introgressed variants as there was with Neanderthals.
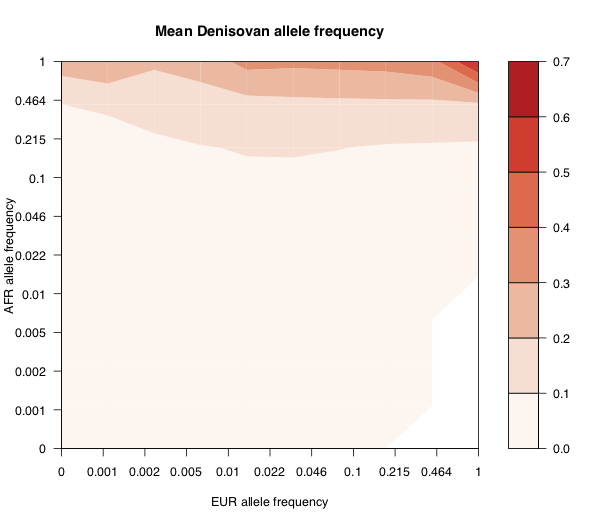
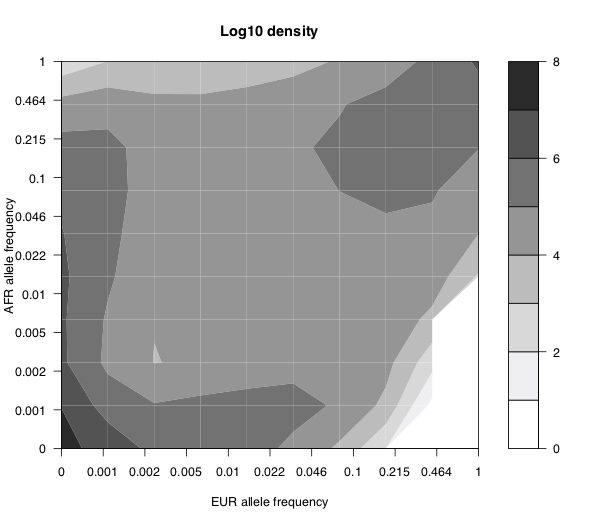
You might think that these patterns are driven in part by repeat mutations. However we see almost the same patterns in C>T/G>A mutations (which you would expect to have higher mutation rates), and non-C>T/G>A mutations, so I do not think that is the main effect. In both of the plots above there’s no colour in the very bottom right corner. That’s because there’s no data there because there are no variants that are very high frequency in Europe and completely absent in Africa. Presumably this is either because there are no Neanderthal-specific variants which swept to high frequency after introgression and/or there is enough gene flow between Africa and non-Africa to bring any such variants to Africa. The plot below shows the relative density of the European-African frequency spectrum.
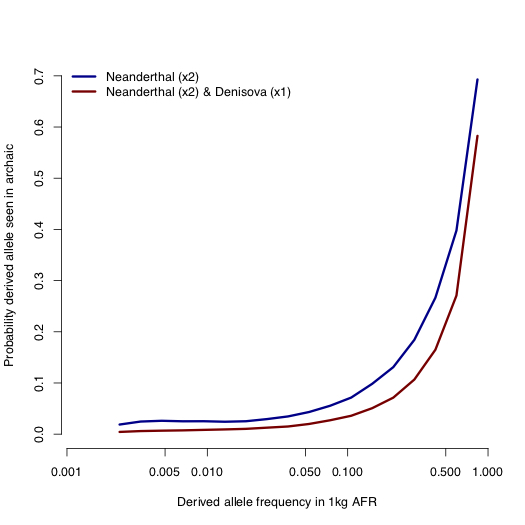
Here’s another way of visualizing the data, showing the probability that an allele at a given frequency in Africa is found (at any frequency) in Neanderthals or, in both Neanderthals and Denisovans.
You can download the processed list here (427Mb). Briefly, I merged the 1000 Genomes phase 3 list of sites with the Altai and Vindija Neanderthal calls, and the Denisovan calls, restricted to sites that had a high-confidence ancestral allele in 1000 Genomes (~47 million sites), and extracted the EUR, EAS, AFR, Denisovan and Neanderthal derived allele frequencies. So this doesn’t include sites that are polymorphic in Neanderthals but not in 1000 Genomes. Please let me know if you find any mistakes!